Deoxyribonucleic Acid (DNA)
Introduction

|
| Image courtesy of freepik.com Opens in new window DNA is a complex double-stranded molecule (called a double helix) in which is coded the information for making amino acids and proteins, essential components of our bodies. Deoxyribonucleic acid or DNA is the ultimate molecule of life composed of four subunits, the bases A, G, C and T, whose order contains instructions for the production of cellular components and for the transmission of identical instructions to daughter cells. |
The instructions that direct human cells to grow, to differentiate into specialized structures, to divide, and to respond to environmental changes are all encoded within the elegant simplicity of the human DNA genome. And the basis of all genetic disease ultimately is founded on changes in this DNA sequence.
DNA is mainly an extraordinary informational macromolecule that plays two central biological roles:
- it carries the instructions for making the components of a cell (mostly proteins, which themselves can manufacture further components);
- it provides a means for this set of instructions to be passed to the daughter cells when a cell divides.
DNA's Double Helix Structure
An understanding of the structure and behavior of DNA and of the chromosomes into which it is packaged is fundamental to medical genetics Opens in new window and to medicine in general, especially as these fields have moved from descriptive into molecular sciences.
The DNA molecular structure is a double helix, made up of two intertwined chains, running in opposite directions, as depicted schematically in Figure X-1.
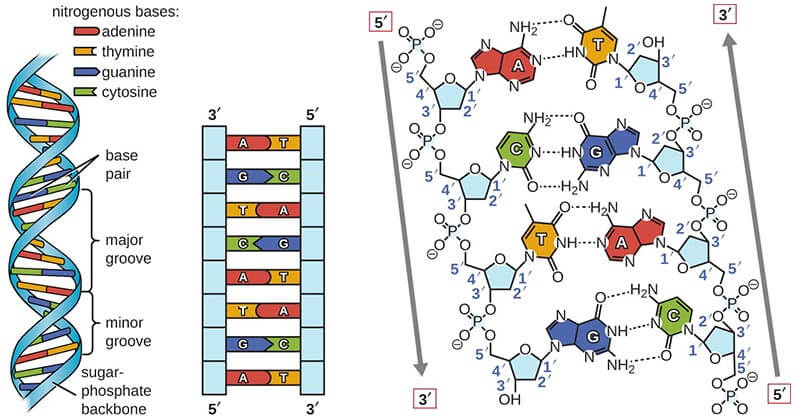
|
| Figure X-1 | The structure of DNA. Adapted from Microbe Notes Opens in new window At the left is a schematic drawing of the DNA double helix, showing the sugar-phosphate backbone as a ribbon, with the bases arranged toward the middle. Note that A always pairs with T, and C always pairs with G. At the right is an expanded view of four nucleotides along one strand, showing the complete chemical structure of the sequence 5’–ACGT–3’. (A nucleotide consists of a sugar, a phosphate group, and an attached base.) Note that the 5’ and 3’ designations, used to indicate the polarity of a DNA strand, refer to the numbering of carbons on the deoxyribose ring. |
The backbone of each strand of the helix consists of an invariant sugar-phosphate polymer, with the sugar being deoxyribose and the phosphates being attached through ester bonds to its 3’ – 5’–hydroxyl groups.
The double helix is held together by hydrogen bonds, which can form between A and T bases and between G and C bases (Figure X-1), each of which is called a base pair.
Thus, the two strands of DNA are complementary; if one is 5’ – ATGCCAG–3’, the other must be 5’–CTGGCAT–3’, with the full double-stranded structure of 7 base pairs (bp) being written as follows:
- 5’–ATGCCAG–3’
- 3’–TACGGTC–5’
There are a number of features of this structure that should be carefully noted:
- Feature #1
It provides a means of storing and coding vast amounts of information, based on the sequence of the bases present in the DNA strand; for a molecule N bases long, there are 4N possible sequences.
The complete DNA sequence of a organism, containing its complete genetic information, is called its genome Opens in new window.
The smallest viruses have genomes of only a few thousand base pairs and contain only a small number of genes Opens in new window.
The size and complexity of the genome increase in a nonlinear fashion, however, as one moves along the evolutionary tree, from 4 x 106 bp in a bacterium to 3 X 109 bp in humans.
A representative sample of genome sizes is shown in Table X-1.
| Table X-1 | Representative Genome Sizes in Base Pairs | |
|---|---|
| SV40, a mammalian DNA virus | 4 x 103 |
| λ, bacterial virus | 5 x 104 |
| Escherichia coli, a bacterium | 4 x 106 |
| Saccharomyces cerevisiae, a yeast | 1.2 x 107 |
| Caennorhabitis elegans, a nematode | 1 x 108 |
| Drosophilia melanogaster, the fruit fly | 1.2 x 108 |
| Human chromosome 21 | 5 x 107 |
| Human chromosome 1 | 3 x 108 |
| Entire human genomea | 3 x 109 |
| Note. This is actually the haploid genome, or half the size of the number of base pairs of DNA in a human somatic cell. The human is a diploid organism, with two copies of each chromosome, except for the sex chromosomes in males. Thus, there are actually 6 x 109 bp of DNA in each human somatic cell. | |
- Feature #2
As noted by Watson and Crick, the double helical complementary structure immediately suggests a mechanism of DNA replication. Each strand contains the full informational content of the DNA molecule and can serve as a template for synthesis of a new complementary strand as the helix unwinds and replicates (depicted in Figure X-2).
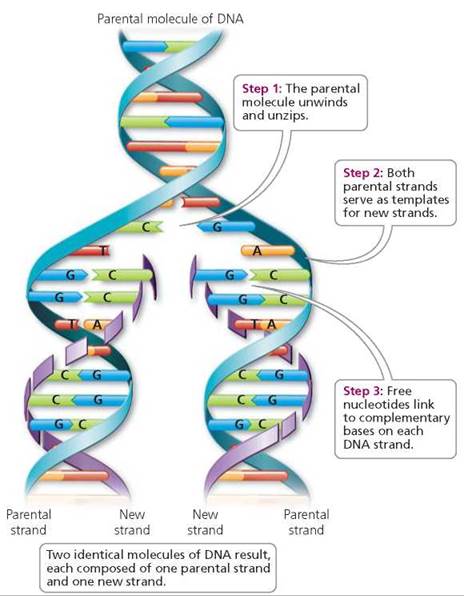
|
| Figure X-2 | Replication of DNA. Identical daughter DNA molecules are generated by unwinding of the parental molecule, with each strand acting as a template for synthesis of a new strand using base pairing rules. |
This mode of replication is denoted semi-conservative because each daughter DNA strand contains one parental strand and one newly synthesized strand.
- Feature #3
The complementary structure also provides a defense against information loss by DNA damage. A base on one strand that is damaged or lost can be replaced using the complementary strand to direct its repair.
Similarly, a break in the sugar-phosphate backbone, which would be nearly impossible to correctly reconnect in a single-stranded molecule, can be repaired in a double-stranded molecule without any loss of contiguity.
- Feature #4
The complementary of DNA strands also allows them to find each other in a complex mixture of molecules. This reannealing or hybridization process is used in some situations by the nuclear machinery to regulate gene expression. Furthermore, this phenomenon has been heavily exploited in molecular biology, and it is at the heart of its current success.
Chemical Structures of the Nucleotide Components of DNA
DNA is a polymer, and its monomeric subunits are molecules called nucleotides. The nucleotide consists in four varieties, which differ in portions known as bases.
The bases are adenine, guanine, cytosine and thymine, abbreviated as A, G, C and T, and it is the sequence of these parts of nucleotide molecules that carries the genetic information. Adenine and guanine, double-ringed molecules, are purines, and cytosine and thymine, single-ringed, are pyrimidines.
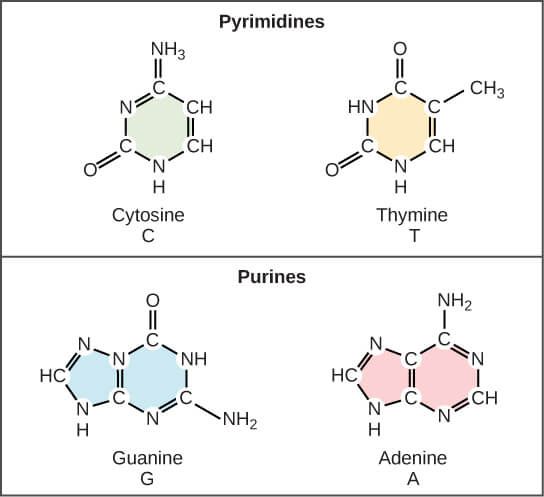
|
| Figure X-3 | Chemical structures of the nucleotide components of DNA. These are the standard four nucleotides; some bases in DNA are modified by the addition of other chemical groups. When nucleotides are incorporated into DNA, two of the phosphate groups are lost. |
Each base joined to a sugar molecule, deoxyribose, and each deoxyribose has a phosphate group attached to it; the sugar and phosphate play only a structural role in DNA, and themselves carry no information. The structures of these nucleotides are shown in Figure X-3.
Some nucleotides can be chemically modified after their incorporation into DNA: an important example is the methylation of cytosine, to give 5-methylcytosine. Methylation plays a role in gene regulation.
The phosphate of one nucleotide is joined to the sugar of another (depicted in Figure X-4b), and so on, and this forms the sugar phosphate backbone of DNA.
Because the deoxyribose group itself is asymmetrical, it provides a polarity to the backbone. The carbon atoms making up the deoxyribose molecule are given numbers from 1’ (pronounced ‘one prime’) to 5’.
Phosphate groups attach to the 3’ and 5’ carbon atoms of the ring, and this provides a way to refer to the different ends of a DNA molecule: the 3’ end has a free hydroxyl (–OH) group (unattached to another nucleotide) on the 3’ carbon, and the 5’ end has a free hydroxyl group on the 5’ carbon.
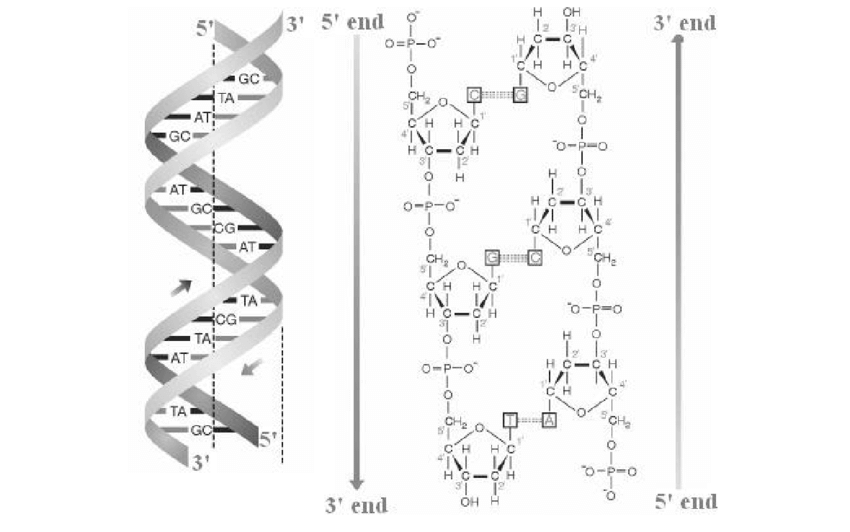
|
| Figure X-4 | Double-stranded helical structure of DNA. (a) The two strands are wound round each other in a double helix. Arrows indicate the direction of each strand, 5’ to 3’. (b) The two strands of DNA are anti-parallel since the linking of the 3’ carbon atom of one base to the 5’ carbon of the next is in opposite directions on the two strands. Since sequences are written 5’ to 3’, the sequence of this three-base DNA molecule on the left-hand strand is CGT, while on the right-hand strand is ACG. |
The polarity of a nucleic acid molecule [whether DNA or RNA (ribonucleic acid)] is important because fundamental processes like DNA replication, the transcription of DNA into RNA, and the translation of RNA into protein will proceed in one direction only along its length. By convention, the sequence of bases in a DNA or RNA molecule is always written left to right in the direction 5’ to 3’.
The structure described above is that of single-stranded DNA.
Within a cell, DNA exists for most of the time in the double-stranded form, as two long strands, spiraling round each other in a double helix (Figure X-4a). The bases of one strand project into the core of the helix, and here they pair with the bases of the other, complementary, strand.
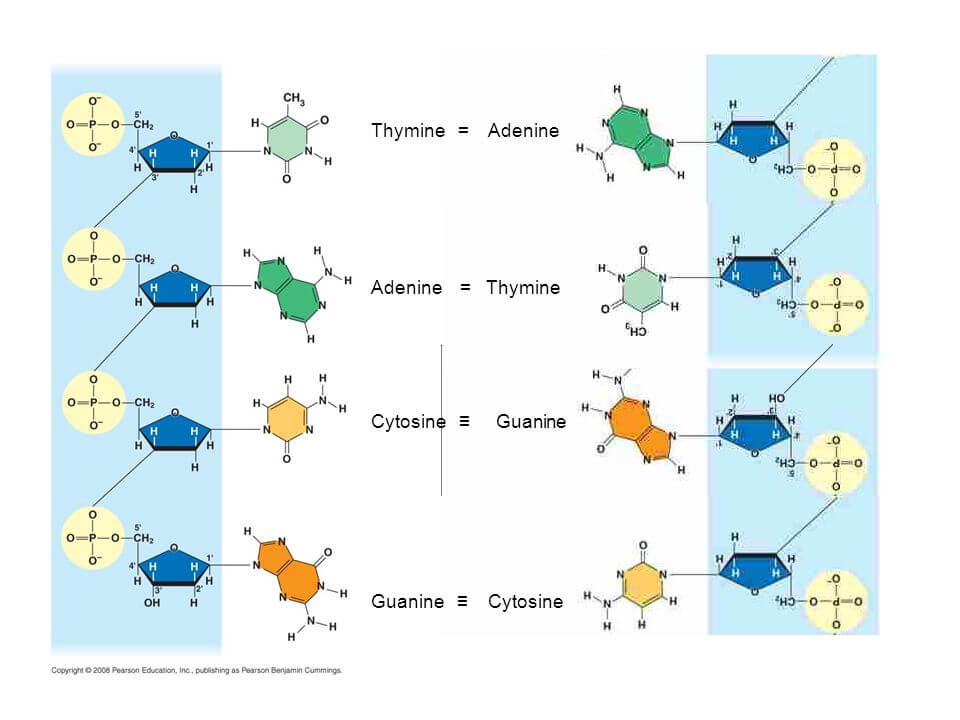
|
| Figure X-5 | Base-pairing between thymine and adenine, and between cytosine and guanine. |
Base-pairing (Figure X-5), where an A pairs strictly with a T, and a C with a G, underlies the two fundamental roles of DNA expression of genetic information via RNA, and replication of genetic information prior to cell division.
A single strand of DNA can act as a template either for the enzymatic synthesis of a complementary strand of RNA (transcription), using ribonucleotides as building blocks, or of a complementary DNA strand (replication), when the building blocks are deoxyribonucleotides.
The lengths of double-stranded DNA molecules are described in units of base-pairs (bp), and for longer molecules kilobase-pairs (usually abbreviated to kilobases, kb) or megabase-pairs (megabases, Mb).
The chemical bonds that tie the atoms together within a single strand of DNA are strong and require considerable energy to break them — these are covalent bonds.
The hydrogen bonds which exist between the bases of one strand and another within a double helix of DNA are much weaker, and can be broken in vitro by relatively gentle processes such as brief heating to 95oC or exposure to alkaline pH, or by active processes within cells.
This separation of the strands of a double helix is called denaturation, or ‘melting’, and must occur prior to either transcription or replication. There are three hydrogen bonds between a G and a C, and two between an A and a T; more energy is needed to denature GC-rich DNA than an equivalent length of AT-rich DNA.
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. 3rd ed. New York: Garland, 1994.
- Darnell J, Lodish H, Baltimore D. Molecular cell biology. 2nd ed. New York: Scientific American Books, WH Freeman, 1990.
- Watson JD, Tooze J, Kurtz DT. Recombinant DNA, a short course. New York: Scientific American Books, WH Freeman, 1983.
- Watson JD, Hopkins NH, Roberts JW, Steitz JA, Weiner AM. Molecular biology of the gene. 4th ed. Menlo Park, CA: Benjamin/Cummings Publishing Co., 1987.
- Meselson M, Stahl FW. The replication of DNA in E. coli Proc Natl Acad Sci USA 1958;44-671-682.

