Ribonucleic Acid (RNA)
Introduction
|
Ribonucleic Acid (RNA) is a polymeric molecule essential in various biological processes within the cell in coding, decoding, regulation and expression of genes. The main function of RNA is to carry information of amino acid sequence from the genes to where proteins are assembled. |
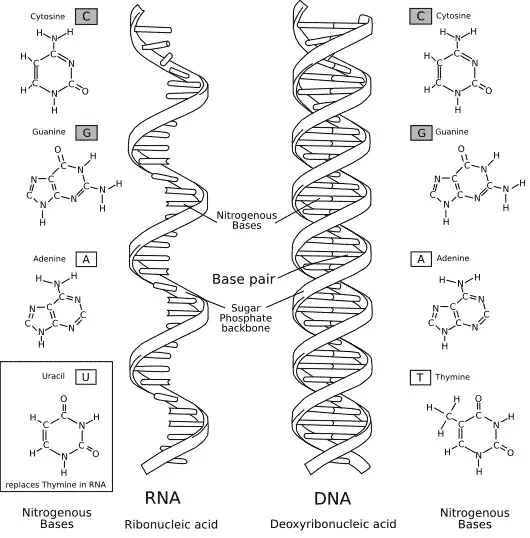
The composition of RNA is very similar to DNA Opens in new window (see Figure X-1), and it plays a key role in all stages of gene expression Opens in new window. RNA is also a linear polynucleotide, but it differs from DNA in that it is single stranded and composed of polymers of ribose rather than deoxyribose, the pyrimidine base uracil (U) replaces thymine (T), and it is relatively unstable when compared to DNA.
|
| Figure X-1 | Comparison of the chemical structures of DNA and RNA. Illustration courtesy of Diffen Opens in new window. The DNA sequence would be written 5’–GT–3’; the RNA sequence would be written 5’–GU–3’. The DNA base thymine is replaced in RNA by uracil, which lacks the methyl group on the pyrimidine ring. Shown also is the additional difference that DNA is ordinarily double-stranded whereas RNA is ordinarily single-stranded. |
There are at least five different types of RNA in eukaryotic cells and all are involved in gene expression:
- Messenger RNA (mRNA) molecules are long, linear, single-stranded polynucleotides that are direct copies of DNA, mRNA is formed by transcription of DNA.
- Small nuclear RNA (snRNA) is a short, ±150 nucleotide (nt) RNA molecule that forms, together with a protein, the small nuclear ribonucleoprotein (snRNP). Several of these snRNPs form with other proteins the splisosoom that facilitates the removal of introns from the precursor mRNA.
- Ribosomal RNA (rRNA) is a structural and functional component of ribosomes. Ribosomes, the cystoplasmic machines that synthesis mRNA into amino acid polypeptides are present in the cytoplasm of the cell and are composed of rRNA and ribosomal proteins.
- Transfer RNA (tRNA) is a small RNA molecule that donates amino acids during translation or protein synthesis.
- A group of small RNAs such as microRNAs (miRNA), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs) and repeat-associated siRNAs (rasiRNAs). These small RNA molecules have various functions at the transcriptional and/or post-transcriptional level, including the break-down of mRNA, repression of transcription and chromatin remodeling.
RNA is about 10 times more abundant than DNA Opens in new window in eukaryotic cells; 80% is rRNA, 15% is tRNA and 5% is mRNA. In the cell mRNA is normally found associated with protein complexes called messenger ribonucleoprotein (mRNP), which package mRNA and aid its transport into the cytoplasm, where it is decoded into a protein.
Transcription
RNA transcription, which is the process whereby the genetic information encoded in DNA is transferred into mRNA, is the first step in the process of gene expression and occurs in the nucleus of the cell.
RNA transcription can be divided into four stages:
- template recognition,
- initiation,
- elongation and
- termination.
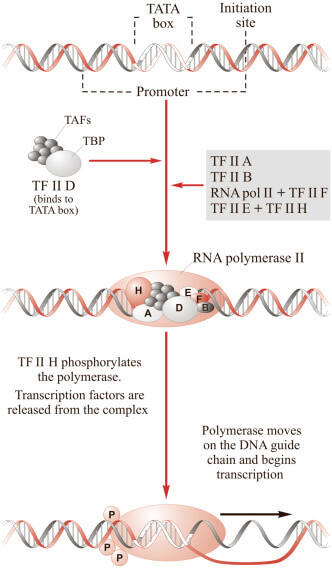
Transcription is catalyzed by DNA-dependent RNA polymerase enzymes (depicted in Figure X-2). In eukaryotic cells RNA polymerase II synthesizes mRNA, while RNA polymerase I and RNA polymerase III synthesize tRNA and rRNA.
RNA polymerase II is a complex 12-subunit enzyme and is associated with several transcription factors (TFs), including TFIIA, TFIIB, TFIIC, TFIID, TFIIE, TFIIE, TFIIH and TFIIJ.
|
| Figure X-2 | RNA polymerase II transcribes the information in DNA into RNA. |
The strand of DNA that directs synthesis of mRNA via complementary base pairing is called the template or antisense strand. The other DNA strand that bears the same nucleotide sequence is called the coding or sense strand. Therefore, RNA represents a copy of DNA Opens in new window.
Transcription is a multistep process, initiated by the assembly of an initiation complex (TFIID, TFIIA, TFIIB, TFIIF, TFIIE, TFIIH) at the promoter, to which RNA polymerase II binds.
TFIID recognizes the promoter and binds to the TATA box at the start of the gene Opens in new window. TFIIH unwinds double-stranded DNA to expose the unpaired DNA nucleotide and the DNA sequence is used as a template from which RNA is synthesized. TFIIE and TFIIH are required for promoter clearance, allowing RNA polymerase II to commence movement away from the promoter.
Initiation describes the synthesis of the first nine nucleotides of the RNA transcript. Elongation describes the phase during which RNA polymerase moves along the DNA and extends the growing RNA molecule.
The RNA molecule is synthesized by adding nucleotides to the free 3’-OH end of the growing RNA chain.
As only at this free 3’-OH end new nucleotides can be attached, RNA synthesis always takes place in the 5’-3’ direction. Growing of the RNA chain is ended by a process called termination, which involves recognition of the terminator sequence that signals the dissociation of the polymerase complex.
Post-transcriptional Process of RNA
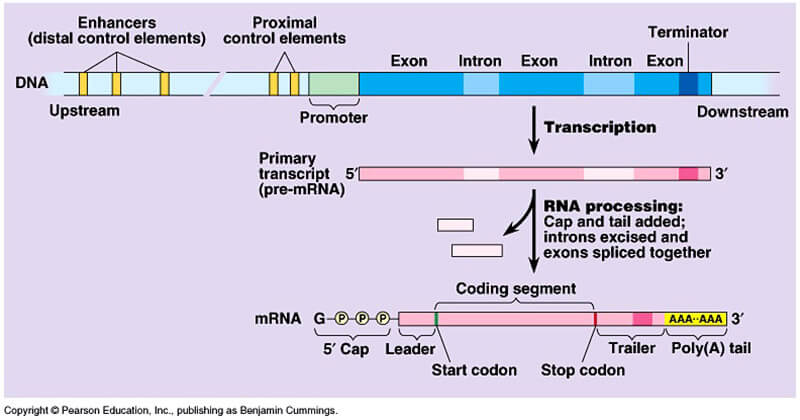
After transcription of DNA into RNA, the newly synthesized RNA is modified. This process is called post-transcriptional processing of RNA (Figure X-3).
|
| Figure X-3 | Transcription and processing of mRNA. |
The primary RNA transcript synthesized by RNA polymerase II from genomic DNA is often called heterogeneous nuclear RNA (hnRNA) because of its considerable variation in size. Instead of hnRNA often the term precursor mRNA (pre-mRNA) is used to designate that it refers to unprocessed RNA.
The primary transcript is an RNA molecule that represents a full copy of the gene extending from the promoter to the terminator region of the gene, and includes introns and exons.
While still in the nucleus, the newly synthesized RNA is capped, polyadenylated and spliced.
Capping refers to the addition of a modified guanine (G) nucleotide (7-methylguanosine) at the 5’ end of the mRNA. This 7-methylguanosine cap has three functions:
- it protects the synthesized RNA from enzymic attack,
- it aids pre-mRNA splicing and
- it enhances translation of the mRNA.
Polyadenylation involves the addition of a string of adenosine (A) residues (a poly-A tail) to the 3’ end of the pre-mRNA. Then the pre-mRNA is spliced, which is performed by the splicosome.
This RNA-protein complex recognizes the consensus sequences at each end of the intron (5’-GU and AG-3’) and excises the introns so that the remaining exons are spliced together to form the mature mRNA molecule. After capping, polyadenylation and splicing, the mRNA is transported from the nucleus to the cytoplasm for translation.
Alternative splicing describes the process whereby other, alternative, splice recognition sites leads to the removal of certain exons, thereby generating different mature mRNA molecules and ultimately different proteins. One primary RNA transcript can be spliced in three different ways, leading to three mRNA isoforms, one with five exons or two with four exons.
Upon translation the different mRNA isoforms will give rise to different isoforms of the protein product of the gene. This is a relatively common phenomenon and although the physiological or metabolic relevance of the different isoforms of many proteins is not fully understood it may be relevant to molecular nutrition.
For example, the peroxisome proliferator activator receptor-gamma (PPARγ) gene can produce three different isoforms of mRNA (PPARγ1, PPARγ2, PPARγ3) as a result of different promoters and alternative splicing.
It would seem that the PPARγ2 mRNA isoform is responsive to nutritional states. PPARγ2 mRNA expression is increased in the fed state; its expression is positively related to adiposity and is reduced on weight loss. Therefore, PPARγ2 mRNA may be the isoform that mediates nutrient regulation of gene expression.
Besides alternative splicing, a peculiar way of splicing exists that occurs with introns that form a ribozyme. These special introns are RNA molecules that function as enzymes and catalyse their own excision, which is referred to as self-splicing.
RNA editing is another way in which the primary RNA transcript can be modified. Editing involves the binding of proteins or short RNA templates to specific regions of the primary transcript and subsequent alteration or editing of the RNA sequence, either by insertion of one or more different nucleotides or by base changes.
This mechanism allows the production of different proteins from a single gene under different physiological conditions. The two isoforms of apo B are examples of RNA editing.
In this case, RNA editing allows for tissue-specific expression of the apo B gene, such that the full protein apo B100 is produced in the liver and secreted on VLDL.
By contrast, in the intestine the apo B48 isoform is produced because a premature stop codon is inserted sot that transcription of the protein is halted and the resultant apoprotein is only 48% of that of the full-length apo B100 protein.
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular biology of the cell. 3rd ed. New York: Garland, 1994.
- Darnell J, Lodish H, Baltimore D. Molecular cell biology. 2nd ed. New York: Scientific American Books, WH Freeman, 1990.

